from IPython.display import Image
from IPython.core.display import HTML
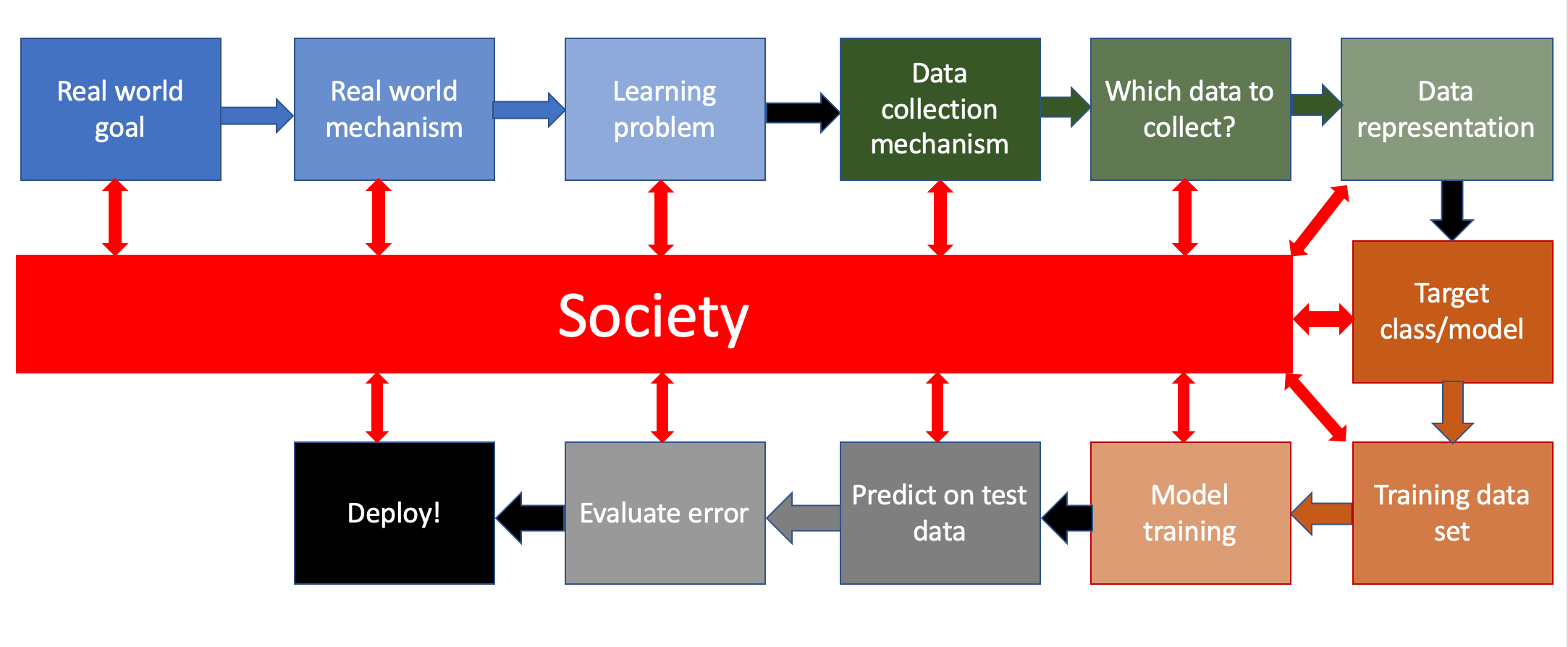
Image(url= "http://www-student.cse.buffalo.edu/~atri/algo-and-society/support/notes/pipeline/images/ML-pipeline-duame-soc.png",
width=800)
import numpy as np
import pandas as pd
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import StandardScaler, OneHotEncoder, PolynomialFeatures
from sklearn.datasets import make_classification
from sklearn.pipeline import Pipeline, make_pipeline
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import validation_curve, learning_curve
We're going to take a look at data on COVID coverage by local news agencies in the US.
There are a number of features in this dataset, there's a more detailed description of most of them here. I'll describe what we're using here, though, obviusly
Note: I am, again, using data that I have used for research in the past. I think it is useful, because I am able to provide more thoughtful responses to questions and comments about the data and methods. It is not because I think my research is especially great, or that you should read it.
covid_df = pd.read_csv("covid_local_news.csv")
from IPython.display import Image
from IPython.core.display import HTML
Image(url= "http://www-student.cse.buffalo.edu/~atri/algo-and-society/support/notes/pipeline/images/ML-pipeline-duame-soc.png",
width=800)

Find local news deserts. In other words, find places where people aren('t) likely to be getting adequate news about COVID
Identify places where the number of articles that cover COVID is low. We can then try to forecast coverage in locations where we don't have data.
from IPython.display import Image
from IPython.core.display import HTML
Image(url= "./coverage.png", width=600, height=600)

px.histogram(covid_df, x="n_covid_full_filter")
px.histogram(covid_df, x="perc_full")
covid_df['log_odds_of_covid_article'] = np.log((covid_df.n_covid_full_filter+1)/(covid_df.n_total_articles-covid_df.n_covid_full_filter+1))
px.histogram(covid_df, x="log_odds_of_covid_article")
Predict the percentage of weekly coverage devoted to COVID for local news outlets across the country that make data available
Below, we display the first five rows of the data...
pd.set_option('display.max_columns', None)
covid_df.head()
| fips_full | fips | state | week_num | fips_covid | sourcedomain_id | lon | lat | title | rank | n_total_articles | n_covid_limited_filter | n_covid_full_filter | fips_cum_cases | fips_cum_deaths | fips_lag1_cum_cases | fips_lag2_cum_cases | fips_n_cases | fips_lag1_n_cases | fips_lag1_cum_deaths | fips_lag2_cum_deaths | fips_n_deaths | fips_lag1_n_deaths | state_cum_cases | state_cum_deaths | state_lag1_cum_cases | state_lag2_cum_cases | state_n_cases | state_lag1_n_cases | state_lag1_cum_deaths | state_lag2_cum_deaths | state_n_deaths | state_lag1_n_deaths | country | country_cum_cases | country_cum_deaths | country_lag1_cum_cases | country_lag2_cum_cases | country_n_cases | country_lag1_n_cases | country_lag1_cum_deaths | country_lag2_cum_deaths | country_n_deaths | country_lag1_n_deaths | date | county | trump16 | clinton16 | total_population | white_pct | black_pct | hispanic_pct | age65andolder_pct | lesshs_pct | rural_pct | ruralurban_cc | state_popn | POPESTIMATE2019 | country_population | lo_trump_vote_16 | fips_n_cases_per1k | fips_n_cases_per1k_log | fips_n_cases_lo | fips_lag1_n_cases_per1k | fips_lag1_n_cases_per1k_log | fips_lag1_n_cases_lo | fips_n_deaths_per1k | fips_n_deaths_per1k_log | fips_n_deaths_lo | fips_lag1_n_deaths_per1k | fips_lag1_n_deaths_per1k_log | fips_lag1_n_deaths_lo | state_n_cases_per1k | state_n_cases_per1k_log | state_n_cases_lo | state_lag1_n_cases_per1k | state_lag1_n_cases_per1k_log | state_lag1_n_cases_lo | state_n_deaths_per1k | state_n_deaths_per1k_log | state_n_deaths_lo | state_lag1_n_deaths_per1k | state_lag1_n_deaths_per1k_log | state_lag1_n_deaths_lo | country_n_cases_per1k | country_n_cases_per1k_log | country_n_cases_lo | country_lag1_n_cases_per1k | country_lag1_n_cases_per1k_log | country_lag1_n_cases_lo | country_n_deaths_per1k | country_n_deaths_per1k_log | country_n_deaths_lo | country_lag1_n_deaths_per1k | country_lag1_n_deaths_per1k_log | country_lag1_n_deaths_lo | perc_full | perc_lim | predrt_0 | predrt_3 | predrt_12 | popuni | log_pop | log_rank | fips5 | Biden | Trump | lo_trump_vote | log_odds_of_covid_article | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1013 | 1013 | Alabama | 14 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 4 | 2 | 4 | 8 | 0 | 2 | 1 | 6 | 1 | 0 | 0 | 0 | 0 | 3955 | 115 | 2200 | 1008 | 1755 | 1192 | 64 | 14 | 51 | 50 | USA | 604363 | 29145 | 393848 | 182876 | 210515 | 210972 | 14501 | 3974 | 14644 | 10527 | 2020-04-08 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 30.851501 | 3.461085 | -7.929641 | 5.141917 | 1.815137 | -9.182404 | 0.000000 | 0.000000 | -9.875551 | 0.000000 | 0.000000 | -9.875551 | 17.896531 | 2.938978 | -8.627749 | 12.155364 | 2.576830 | -9.014316 | 0.520070 | 0.418756 | -12.147299 | 0.509873 | 0.412025 | -12.166717 | 32.067284 | 3.498544 | -8.045084 | 32.136898 | 3.500647 | -8.042916 | 2.230688 | 1.172695 | -10.710547 | 1.603555 | 0.956878 | -11.040608 | 1.000000 | 1.000000 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 | 1.609438 |
| 1 | 1013 | 1013 | Alabama | 15 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 11 | 1 | 7 | 16 | 0 | 8 | 2 | 8 | 6 | 0 | 0 | 0 | 0 | 5350 | 195 | 3955 | 2200 | 1395 | 1755 | 115 | 64 | 80 | 51 | USA | 804845 | 44373 | 604363 | 393848 | 200482 | 210515 | 29145 | 14501 | 15228 | 14644 | 2020-04-15 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 41.135335 | 3.740887 | -7.678326 | 30.851501 | 3.461085 | -7.929641 | 0.000000 | 0.000000 | -9.875551 | 0.000000 | 0.000000 | -9.875551 | 14.225447 | 2.722968 | -8.857177 | 17.896531 | 2.938978 | -8.627749 | 0.815796 | 0.596524 | -11.704094 | 0.520070 | 0.418756 | -12.147299 | 30.538979 | 3.451224 | -8.093917 | 32.067284 | 3.498544 | -8.045084 | 2.319648 | 1.199859 | -10.671445 | 2.230688 | 1.172695 | -10.710547 | 0.636364 | 0.636364 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 | 0.470004 |
| 2 | 1013 | 1013 | Alabama | 16 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 1 | 0 | 1 | 45 | 1 | 16 | 8 | 29 | 8 | 0 | 0 | 1 | 0 | 6752 | 245 | 5350 | 3955 | 1402 | 1395 | 195 | 115 | 50 | 80 | USA | 1012453 | 58064 | 804845 | 604363 | 207608 | 200482 | 44373 | 29145 | 13691 | 15228 | 2020-04-22 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 149.115590 | 5.011406 | -6.474354 | 41.135335 | 3.740887 | -7.678326 | 5.141917 | 1.815137 | -9.182404 | 0.000000 | 0.000000 | -9.875551 | 14.296830 | 2.727646 | -8.852175 | 14.225447 | 2.722968 | -8.857177 | 0.509873 | 0.412025 | -12.166717 | 0.815796 | 0.596524 | -11.704094 | 31.624467 | 3.485063 | -8.058990 | 30.538979 | 3.451224 | -8.093917 | 2.085520 | 1.126720 | -10.777834 | 2.319648 | 1.199859 | -10.671445 | 1.000000 | 1.000000 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 | 0.693147 |
| 3 | 1013 | 1013 | Alabama | 17 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 12 | 3 | 7 | 120 | 2 | 45 | 16 | 75 | 29 | 1 | 0 | 1 | 1 | 8438 | 317 | 6752 | 5350 | 1686 | 1402 | 245 | 195 | 72 | 50 | USA | 1203928 | 70834 | 1012453 | 804845 | 191475 | 207608 | 58064 | 44373 | 12770 | 13691 | 2020-04-29 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 385.643768 | 5.957504 | -5.544818 | 149.115590 | 5.011406 | -6.474354 | 5.141917 | 1.815137 | -9.182404 | 5.141917 | 1.815137 | -9.182404 | 17.192906 | 2.901032 | -8.667836 | 14.296830 | 2.727646 | -8.852175 | 0.734217 | 0.550556 | -11.808083 | 0.509873 | 0.412025 | -12.166717 | 29.166963 | 3.406747 | -8.139884 | 31.624467 | 3.485063 | -8.058990 | 1.945226 | 1.080185 | -10.847469 | 2.085520 | 1.126720 | -10.777834 | 0.583333 | 0.583333 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 | 0.287682 |
| 4 | 1013 | 1013 | Alabama | 18 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 3 | 1 | 3 | 224 | 6 | 120 | 45 | 104 | 75 | 2 | 1 | 4 | 1 | 10464 | 436 | 8438 | 6752 | 2026 | 1686 | 317 | 245 | 119 | 72 | USA | 1369338 | 82081 | 1203928 | 1012453 | 165410 | 191475 | 70834 | 58064 | 11247 | 12770 | 2020-05-06 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 534.759358 | 6.283685 | -5.221591 | 385.643768 | 5.957504 | -5.544818 | 20.567668 | 3.071195 | -8.266113 | 5.141917 | 1.815137 | -9.182404 | 20.660040 | 3.075469 | -8.484231 | 17.192906 | 2.901032 | -8.667836 | 1.213497 | 0.794574 | -11.311051 | 0.734217 | 0.550556 | -11.808083 | 25.196539 | 3.265627 | -8.286213 | 29.166963 | 3.406747 | -8.139884 | 1.713231 | 0.998140 | -10.974456 | 1.945226 | 1.080185 | -10.847469 | 1.000000 | 1.000000 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 | 1.386294 |
In general, here, we would also work to clean up our data for analysis - rescaling features, dropping features, one-hot encoding, etc. For the purposes of this notebook, though, where we're going to do some exploration, I'm going to leave that to the modeling section
We're going to stick with linear regression for now, but we can choose to add noon-linear . Now we know what that is! And how to optimize it (although we'll use the sklearn implementation
Next week, we'll cover model evaluation in more detail. For now, we're just going to note that to ensure our model is generalizable and not overfit to the training data, we need to separate out a training dataset from a test dataset.
Exercise: Give a high-level argument for why evaluating on the training data is a bad idea
With temporal data, and more generally, data with dependencies, it is also important that we make sure that we are avoiding the leakage of information from the training data in a way that creates a biased understanding of how well we are making predictions. Leakage can happen in at least two ways (again, we'll go into more detail next week):
For this simple example, for now, we're going to ignore the leakage issue. We'll come back and fix that next week
Neat! But to pick a good training dataset, we first need to know ... what our dataset is. This data has a lot of features. In class, we'll play with a bunch of them, together. Here, I'm just going to get us started
CONTINUOUS_FEATURES = ["state_lag1_n_cases_per1k",
"fips_lag1_n_cases_per1k",
"country_lag1_n_cases_per1k",
"predrt_0"]
OUTCOME = 'log_odds_of_covid_article'
X = covid_df[CONTINUOUS_FEATURES]
y = covid_df[OUTCOME]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=5)
OK, let's have at it!
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train,y_train)
LinearRegression()
predictions = model.predict(X_test)
from sklearn.metrics import mean_squared_error
mean_squared_error(y_test,predictions)
0.6643640375053314
What might we be asking ourselves before we deploy? What might we try to change? Let's work on it!
In class exercise ... beat Kenny's predictive model!
The above pipeline is a barebones representation. In reality, we are trying a bunch of different models, data representations, and even questions, before we deploy. Here's a representation of the pipeline that trends further towards that, from this tweet.
Image(url="https://pbs.twimg.com/media/FKtEyl5XEAABdEa?format=jpg&name=4096x4096", width=800)

px.scatter(covid_df.sample(10000),
x='fips_n_cases_per1k_log',
y='log_odds_of_covid_article',
opacity=.5
)
px.scatter(covid_df.sample(10000),
x='country_n_deaths_per1k',
y='log_odds_of_covid_article',
opacity=.5
)
from scipy.stats import pearsonr
pearsonr(covid_df.country_n_deaths_per1k, covid_df.log_odds_of_covid_article )
(-0.03008625943248823, 0.0004817002592877463)
pearsonr(covid_df.fips_n_deaths_per1k, covid_df.log_odds_of_covid_article )
(-0.009869673637258653, 0.2522552434237165)
averaged_data = covid_df.groupby("date").agg(
cases = pd.NamedAgg("country_n_cases_per1k","first"),
deaths = pd.NamedAgg("country_n_deaths_per1k","first"),
article_odds = pd.NamedAgg("log_odds_of_covid_article","mean")
).reset_index()
fig = px.line(pd.melt(averaged_data,id_vars="date"),x="date",y="value",facet_col="variable")
fig.update_yaxes(matches=None)
fig.show()
One other form of EDA that I personally often leverage is to fit a simple (usually linear) model to the data and explore the coefficients for indicators of what effects might exist.
As we have now discussed in class, thouhg, we have to be really careful when interpreting coefficients for models with transformed predictor variables. Here, for example, is a useful resource for your programming assignment.
In our case, we actually ended up using a variable that means interpreting our coefficients using interpretations for logistic regression. Here is a good explanation. We will cover this in more detail next week, but a simple plot below to discuss!
Exercise: Are the estimates from the coefficients we used comparable? Which are, and which are not? What might we do to make them even more comparable?
For this demo, I took code from this sklearn tutorial. The tutorial is very nice, I would highly recommend going through it, although I will teach most of what is in this over the next week or two in one way or another.
feature_names = CONTINUOUS_FEATURES
coefs = pd.DataFrame(
model.coef_,
columns=["Coefficients"],
index=feature_names,
)
coefs
| Coefficients | |
|---|---|
| state_lag1_n_cases_per1k | 0.000557 |
| fips_lag1_n_cases_per1k | -0.000172 |
| country_lag1_n_cases_per1k | -0.002770 |
| predrt_0 | -0.013255 |
import matplotlib.pyplot as plt
coefs.plot(kind="barh", figsize=(9, 7))
plt.title("Ridge model, small regularization")
plt.axvline(x=0, color=".5")
plt.subplots_adjust(left=0.3)
coefs['odds_ratio'] = np.exp(coefs.Coefficients)
coefs
| Coefficients | odds_ratio | |
|---|---|---|
| state_lag1_n_cases_per1k | 0.000557 | 1.000557 |
| fips_lag1_n_cases_per1k | -0.000172 | 0.999828 |
| country_lag1_n_cases_per1k | -0.002770 | 0.997234 |
| predrt_0 | -0.013255 | 0.986832 |
Some of the other things we've discussed in class don't fit neatly within a real-world ML pipeline, but are useful to show on this dataset... these are now in this section below
from sklearn.model_selection import cross_val_score
%timeit cross_val_score(model, X_train, y_train, cv=10)
%timeit cross_val_score(model, X_train, y_train, cv=100)
303 ms ± 39.8 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
CONTINUOUS_FEATURES = ["state_lag1_n_cases_per1k",
"fips_lag1_n_cases_per1k",
"country_lag1_n_cases_per1k",
"predrt_0", "week_num", "lo_trump_vote"
]
CATEGORICAL_VARS = ['state']
OUTCOME = 'log_odds_of_covid_article'
covid_df = covid_df.sample(frac=1, random_state=1)
X = covid_df[CONTINUOUS_FEATURES +CATEGORICAL_VARS]
y = covid_df[OUTCOME]
basic_pipeline = make_pipeline(
ColumnTransformer([('numerical', StandardScaler(), CONTINUOUS_FEATURES),
("categorical", OneHotEncoder(handle_unknown = 'ignore'),
CATEGORICAL_VARS)]),
LinearRegression()
)
train_sizes, train_scores, valid_scores = learning_curve(
basic_pipeline, X, y,
cv=5,
scoring="neg_root_mean_squared_error",
train_sizes=np.linspace(0.05, 1.0, 20),
random_state=104
)
def gen_curve(scores,curvetype,train_sizes,nfolds,nticks):
return (pd.DataFrame(scores,
columns = [f"fold_{i}" for i in range(nfolds)])
.assign(size=train_sizes)
.assign(curve=[curvetype]*nticks)
)
learning_curve_data = pd.concat(
[gen_curve(train_scores,"Training",train_sizes,5,20),
gen_curve(valid_scores,"Validation",train_sizes,5,20)
]
)
learning_curve_plot_data = (pd.melt(learning_curve_data,
id_vars=['size','curve'])
.groupby(['size','curve'])
.agg(
mean=pd.NamedAgg(column="value", aggfunc=np.mean),
sd=pd.NamedAgg(column="value", aggfunc=np.std)
)
.reset_index()
)
fig = px.scatter(learning_curve_plot_data,
x="size", y="mean", color="curve",
error_y="sd",
labels={
"size":"Size of Training Data",
"mean" : "Negative MSE"
},
height=500,
width=600)
fig.show()
np.random.seed(0)
X = np.sort(5 * np.random.rand(40, 1), axis=0)
T = np.linspace(0, 5, 500)[:, np.newaxis]
y = np.sin(X).ravel()
# Add noise to targets
y[::5] += 1 * (0.5 - np.random.rand(8))
# Generate sample data
import numpy as np
import matplotlib.pyplot as plt
from sklearn import neighbors
cdf = covid_df
X = cdf.week_num[:, np.newaxis]
y = cdf[OUTCOME].values
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=5)
srt = list(np.argsort(X_train,axis=0).ravel())
X_train = X_train[srt]
y_train = y_train[srt]
X_test = np.sort(X_test,axis=0)
# #############################################################################
# Fit regression model
n_neighbors = 1
for i, weights in enumerate(["uniform", "distance"]):
knn = neighbors.KNeighborsRegressor(n_neighbors, weights=weights)
y_ = knn.fit(X_train, y_train).predict(X_test)
plt.subplot(2, 1, i + 1)
plt.scatter(X_train, y_train, color="darkorange", label="data")
plt.plot(X_test, y_, color="navy", label="prediction")
plt.axis("tight")
plt.legend()
plt.title("KNeighborsRegressor (k = %i, weights = '%s')" % (n_neighbors, weights))
plt.tight_layout()
plt.show()
/var/folders/rf/4jz25sh1733_8b28phhhrlfh0000gn/T/ipykernel_12153/3751386291.py:7: FutureWarning: Support for multi-dimensional indexing (e.g. `obj[:, None]`) is deprecated and will be removed in a future version. Convert to a numpy array before indexing instead.
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import load_digits
from sklearn.svm import SVC
from sklearn.model_selection import validation_curve
# #########
param_range = range(1,20)
train_scores, test_scores = validation_curve(
neighbors.KNeighborsRegressor(),
X_train,
y_train,
param_name="n_neighbors",
param_range=param_range,
scoring="neg_root_mean_squared_error",
n_jobs=2,
cv=5
)
learning_curve_data = pd.concat(
[gen_curve(train_scores,"Training",param_range,5,19),
gen_curve(test_scores,"Validation",param_range,5, 19)
]
)
learning_curve_plot_data = (pd.melt(learning_curve_data,
id_vars=['size','curve'])
.groupby(['size','curve'])
.agg(
mean=pd.NamedAgg(column="value", aggfunc=np.mean),
sd=pd.NamedAgg(column="value", aggfunc=np.std)
)
.reset_index()
)
fig = px.scatter(learning_curve_plot_data,
x="size", y="mean", color="curve",
error_y="sd",
labels={
"size":"K",
"mean" : "Negative RMSE"
})
fig.show()
Yikes! Compare that to the standard sklearn example.
What gives?
Image("https://github.com/rasbt/stat451-machine-learning-fs20/"+
"raw/9e463e06a49278a3940774ef32def883e85e9cc0/L11/code/nested-cv-image.png")
import numpy as np
from sklearn.model_selection import GridSearchCV,train_test_split,StratifiedKFold,cross_val_score
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KNeighborsRegressor
from sklearn.tree import DecisionTreeRegressor
from sklearn.metrics import mean_squared_error
CONTINUOUS_FEATURES = ["state_lag1_n_cases_per1k",
"fips_lag1_n_cases_per1k",
"country_lag1_n_cases_per1k",
"predrt_0",
"lo_trump_vote",
"black_pct",
"white_pct",
"lon",
"lat",
"week_num"
]
CATEGORICAL_VARS = ['state']
OUTCOME = 'log_odds_of_covid_article'
covid_df = covid_df.sample(frac=1, random_state=1)
X = covid_df[CONTINUOUS_FEATURES +CATEGORICAL_VARS]
y = covid_df[OUTCOME]
X_train, X_test, y_train, y_test = train_test_split(X, y,
test_size=0.2,
random_state=1)
basic_pipeline = make_pipeline(
ColumnTransformer([('numerical', StandardScaler(), CONTINUOUS_FEATURES),
("categorical", OneHotEncoder(handle_unknown = 'ignore'),CATEGORICAL_VARS)]),
)
clf1 = LinearRegression(fit_intercept=True)
clf2 = KNeighborsRegressor()
clf3 = DecisionTreeRegressor(random_state=1)
# Building the pipelines
pipe1 = Pipeline([("pipe",basic_pipeline),
('clf1', clf1)])
pipe2 = Pipeline([("pipe",basic_pipeline),
('clf2', clf2)])
pipe3 = Pipeline([("pipe",basic_pipeline),
('clf3', clf3)])
# Setting up the parameter grids
param_grid1 = [{'clf1__fit_intercept' : [True,False]}]
param_grid2 = [{'clf2__n_neighbors' : [1,2,5,10]}]
param_grid3 = [{'clf3__max_depth': [5,10],
'clf3__criterion': ["squared_error", "absolute_error"]}]
from sklearn.model_selection import KFold
gridcvs = {}
inner_cv = KFold(n_splits=5, shuffle=True, random_state=1)
for pgrid, est, name in zip((param_grid1, param_grid2, param_grid3),
(pipe1, pipe2,pipe3),
('Linear', 'KNN','DT')):
gcv = GridSearchCV(estimator=est,
param_grid=pgrid,
scoring='neg_mean_squared_error',
cv=inner_cv,
verbose=0,
refit=True)
gridcvs[name] = gcv
for name, gs_est in sorted(gridcvs.items()):
print(50 * '-', '\n')
print('Algorithm:', name)
print(' Inner loop:')
outer_scores = []
outer_cv = KFold(n_splits=5, shuffle=True, random_state=1)
for train_idx, valid_idx in outer_cv.split(X_train, y_train):
[]
gridcvs[name].fit(X_train.iloc[train_idx],
y_train.iloc[train_idx]) # run inner loop hyperparam tuning
print('\n Best MSE (avg. of inner test folds) {}'.format(gridcvs[name].best_score_ *-1))
print(' Best parameters:', gridcvs[name].best_params_)
# perf on test fold (valid_idx)
outer_scores.append(gridcvs[name].best_estimator_.score(X_train.iloc[valid_idx], y_train.iloc[valid_idx]))
print(' MSE (on outer test fold) %.2f' % (outer_scores[-1]*1))
print('\n Outer Loop:')
print(' MSE %.2f +/- %.2f' %
(np.mean(outer_scores), np.std(outer_scores)))
--------------------------------------------------
Algorithm: DT
Inner loop:
Best MSE (avg. of inner test folds) 0.3801930576479365
Best parameters: {'clf3__criterion': 'squared_error', 'clf3__max_depth': 10}
MSE (on outer test fold) 0.50
Best MSE (avg. of inner test folds) 0.385387575033809
Best parameters: {'clf3__criterion': 'squared_error', 'clf3__max_depth': 10}
MSE (on outer test fold) 0.48
Best MSE (avg. of inner test folds) 0.37763460990277814
Best parameters: {'clf3__criterion': 'absolute_error', 'clf3__max_depth': 10}
MSE (on outer test fold) 0.46
Best MSE (avg. of inner test folds) 0.3857138446021934
Best parameters: {'clf3__criterion': 'squared_error', 'clf3__max_depth': 10}
MSE (on outer test fold) 0.48
Best MSE (avg. of inner test folds) 0.38646220655549246
Best parameters: {'clf3__criterion': 'squared_error', 'clf3__max_depth': 10}
MSE (on outer test fold) 0.48
Outer Loop:
MSE 0.48 +/- 0.01
--------------------------------------------------
Algorithm: KNN
Inner loop:
Best MSE (avg. of inner test folds) 0.33695377607512256
Best parameters: {'clf2__n_neighbors': 5}
MSE (on outer test fold) 0.56
Best MSE (avg. of inner test folds) 0.3367630581625388
Best parameters: {'clf2__n_neighbors': 5}
MSE (on outer test fold) 0.57
Best MSE (avg. of inner test folds) 0.33663133662807615
Best parameters: {'clf2__n_neighbors': 5}
MSE (on outer test fold) 0.56
Best MSE (avg. of inner test folds) 0.3403680408023601
Best parameters: {'clf2__n_neighbors': 5}
MSE (on outer test fold) 0.54
Best MSE (avg. of inner test folds) 0.3441921519509416
Best parameters: {'clf2__n_neighbors': 5}
MSE (on outer test fold) 0.56
Outer Loop:
MSE 0.56 +/- 0.01
--------------------------------------------------
Algorithm: Linear
Inner loop:
Best MSE (avg. of inner test folds) 0.4601493154975862
Best parameters: {'clf1__fit_intercept': True}
MSE (on outer test fold) 0.38
Best MSE (avg. of inner test folds) 0.45045088806757166
Best parameters: {'clf1__fit_intercept': False}
MSE (on outer test fold) 0.36
Best MSE (avg. of inner test folds) 0.4519535406462786
Best parameters: {'clf1__fit_intercept': True}
MSE (on outer test fold) 0.36
Best MSE (avg. of inner test folds) 0.4611015435489009
Best parameters: {'clf1__fit_intercept': True}
MSE (on outer test fold) 0.39
Best MSE (avg. of inner test folds) 0.4593054421591429
Best parameters: {'clf1__fit_intercept': False}
MSE (on outer test fold) 0.36
Outer Loop:
MSE 0.37 +/- 0.01
gcv_model_select = GridSearchCV(estimator=pipe1,
param_grid=param_grid1,
scoring='neg_mean_squared_error',
cv=inner_cv,
verbose=1,
refit=True)
gcv_model_select.fit(X_train, y_train)
print('Best CV MSE: {}'.format(gcv_model_select.best_score_*-1))
print('Best parameters:', gcv_model_select.best_params_)
Fitting 5 folds for each of 2 candidates, totalling 10 fits
Best CV MSE: 0.4552929660458946
Best parameters: {'clf1__fit_intercept': False}
from sklearn.metrics import mean_squared_error
train_mse = mean_squared_error(y_true=y_train, y_pred=gcv_model_select.predict(X_train))
test_mse = mean_squared_error(y_true=y_test, y_pred=gcv_model_select.predict(X_test))
print('Training MSE: {}'.format(train_mse))
print('Test MSE: {}'.format(test_mse))
Training MSE: 0.45028534654415026 Test MSE: 0.43868299320467713
import plotly.graph_objects as go
data = pd.DataFrame({"true":y_test,"pred":gcv_model_select.predict(X_test), "state": X_test.state})
fig = px.scatter(data,x="true",y="pred",color='state',height=600,width=700)
fig.add_trace(
go.Scatter(x=data['true'], y=data['true'], name="slope", line_shape='linear')
)
fig.show()
for x in covid_df.columns:
print(x)
fips_full fips state week_num fips_covid sourcedomain_id lon lat title rank n_total_articles n_covid_limited_filter n_covid_full_filter fips_cum_cases fips_cum_deaths fips_lag1_cum_cases fips_lag2_cum_cases fips_n_cases fips_lag1_n_cases fips_lag1_cum_deaths fips_lag2_cum_deaths fips_n_deaths fips_lag1_n_deaths state_cum_cases state_cum_deaths state_lag1_cum_cases state_lag2_cum_cases state_n_cases state_lag1_n_cases state_lag1_cum_deaths state_lag2_cum_deaths state_n_deaths state_lag1_n_deaths country country_cum_cases country_cum_deaths country_lag1_cum_cases country_lag2_cum_cases country_n_cases country_lag1_n_cases country_lag1_cum_deaths country_lag2_cum_deaths country_n_deaths country_lag1_n_deaths date county trump16 clinton16 total_population white_pct black_pct hispanic_pct age65andolder_pct lesshs_pct rural_pct ruralurban_cc state_popn POPESTIMATE2019 country_population lo_trump_vote_16 fips_n_cases_per1k fips_n_cases_per1k_log fips_n_cases_lo fips_lag1_n_cases_per1k fips_lag1_n_cases_per1k_log fips_lag1_n_cases_lo fips_n_deaths_per1k fips_n_deaths_per1k_log fips_n_deaths_lo fips_lag1_n_deaths_per1k fips_lag1_n_deaths_per1k_log fips_lag1_n_deaths_lo state_n_cases_per1k state_n_cases_per1k_log state_n_cases_lo state_lag1_n_cases_per1k state_lag1_n_cases_per1k_log state_lag1_n_cases_lo state_n_deaths_per1k state_n_deaths_per1k_log state_n_deaths_lo state_lag1_n_deaths_per1k state_lag1_n_deaths_per1k_log state_lag1_n_deaths_lo country_n_cases_per1k country_n_cases_per1k_log country_n_cases_lo country_lag1_n_cases_per1k country_lag1_n_cases_per1k_log country_lag1_n_cases_lo country_n_deaths_per1k country_n_deaths_per1k_log country_n_deaths_lo country_lag1_n_deaths_per1k country_lag1_n_deaths_per1k_log country_lag1_n_deaths_lo perc_full perc_lim predrt_0 predrt_3 predrt_12 popuni log_pop log_rank fips5 Biden Trump lo_trump_vote log_odds_of_covid_article
# Write your code for Part 1.2 here
from sklearn.linear_model import Ridge
all_continuous_vars = ["week_num", "lon", "lat", "fips_cum_cases",
"white_pct", "black_pct", "hispanic_pct",
"age65andolder_pct", "lesshs_pct", "rural_pct", "ruralurban_cc", "state_popn", "POPESTIMATE2019",
"lo_trump_vote_16",
"fips_n_cases_per1k", "fips_n_cases_per1k_log", "fips_n_cases_lo", "fips_lag1_n_cases_per1k",
"fips_lag1_n_cases_per1k_log", "fips_lag1_n_cases_lo", "fips_n_deaths_per1k", "fips_n_deaths_per1k_log", "fips_n_deaths_lo",
"fips_lag1_n_deaths_per1k", "fips_lag1_n_deaths_per1k_log", "fips_lag1_n_deaths_lo", "state_n_cases_per1k",
"state_n_cases_per1k_log", "state_n_cases_lo", "state_lag1_n_cases_per1k", "state_lag1_n_cases_per1k_log",
"state_lag1_n_cases_lo", "state_n_deaths_per1k", "state_n_deaths_per1k_log", "state_n_deaths_lo", "state_lag1_n_deaths_per1k",
"state_lag1_n_deaths_per1k_log", "state_lag1_n_deaths_lo", "country_n_cases_per1k", "country_n_cases_per1k_log",
"country_n_cases_lo", "country_lag1_n_cases_per1k", "country_lag1_n_cases_per1k_log", "country_lag1_n_cases_lo",
"country_n_deaths_per1k", "country_n_deaths_per1k_log", "country_n_deaths_lo", "country_lag1_n_deaths_per1k",
"country_lag1_n_deaths_per1k_log", "country_lag1_n_deaths_lo",
"predrt_0", "predrt_3", "predrt_12",
"log_pop", "log_rank", "lo_trump_vote"]
from sklearn.linear_model import Lasso
from sklearn.metrics import mean_squared_error
l1_penalties = np.logspace(-3, 3, num=15) # TODO: make the list of lambda values
lasso_output = []
lasso_data = covid_df[all_continuous_vars+["log_odds_of_covid_article"]].dropna()
for l1_penalty in l1_penalties:
print(l1_penalty)
regression_pipeline = make_pipeline(ColumnTransformer([
("scale",StandardScaler(),all_continuous_vars)
]),
Lasso(alpha=l1_penalty, random_state=0)
)
regression_pipeline.fit(lasso_data,lasso_data.log_odds_of_covid_article)
train_rmse = np.sqrt(mean_squared_error(lasso_data.log_odds_of_covid_article,
regression_pipeline.predict(lasso_data)))
# We maintain a list of dictionaries containing our results
lasso_output.append({
'l1_penalty': l1_penalty,
'model': regression_pipeline,
'train_rmse': train_rmse})
lasso_output = pd.DataFrame(lasso_output)
0.001 0.0026826957952797246 0.0071968567300115215 0.019306977288832496 0.0517947467923121 0.13894954943731375 0.3727593720314938 1.0 2.6826957952797246 7.196856730011514 19.306977288832496 51.794746792312125 138.9495494373136 372.7593720314938 1000.0
lasso_output
| l1_penalty | model | train_rmse | |
|---|---|---|---|
| 0 | 0.001000 | (ColumnTransformer(transformers=[('scale', Sta... | 0.680973 |
| 1 | 0.002683 | (ColumnTransformer(transformers=[('scale', Sta... | 0.682676 |
| 2 | 0.007197 | (ColumnTransformer(transformers=[('scale', Sta... | 0.686630 |
| 3 | 0.019307 | (ColumnTransformer(transformers=[('scale', Sta... | 0.691798 |
| 4 | 0.051795 | (ColumnTransformer(transformers=[('scale', Sta... | 0.706008 |
| 5 | 0.138950 | (ColumnTransformer(transformers=[('scale', Sta... | 0.757093 |
| 6 | 0.372759 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 7 | 1.000000 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 8 | 2.682696 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 9 | 7.196857 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 10 | 19.306977 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 11 | 51.794747 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 12 | 138.949549 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 13 | 372.759372 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
| 14 | 1000.000000 | (ColumnTransformer(transformers=[('scale', Sta... | 0.850479 |
from sklearn import set_config
from sklearn.utils import estimator_html_repr
from IPython.core.display import display, HTML
set_config(display='diagram')
mod = lasso_output.model.iloc[2]
mod
Pipeline(steps=[('columntransformer',
ColumnTransformer(transformers=[('scale', StandardScaler(),
['week_num', 'lon', 'lat',
'fips_cum_cases',
'white_pct', 'black_pct',
'hispanic_pct',
'age65andolder_pct',
'lesshs_pct', 'rural_pct',
'ruralurban_cc',
'state_popn',
'POPESTIMATE2019',
'lo_trump_vote_16',
'fips_n_cases_per1k',
'fips_n_cases_per1k_log',
'fips_n_cases_lo',...
'fips_lag1_n_cases_per1k_log',
'fips_lag1_n_cases_lo',
'fips_n_deaths_per1k',
'fips_n_deaths_per1k_log',
'fips_n_deaths_lo',
'fips_lag1_n_deaths_per1k',
'fips_lag1_n_deaths_per1k_log',
'fips_lag1_n_deaths_lo',
'state_n_cases_per1k',
'state_n_cases_per1k_log',
'state_n_cases_lo',
'state_lag1_n_cases_per1k', ...])])),
('lasso', Lasso(alpha=0.0071968567300115215, random_state=0))])Please rerun this cell to show the HTML repr or trust the notebook.Pipeline(steps=[('columntransformer',
ColumnTransformer(transformers=[('scale', StandardScaler(),
['week_num', 'lon', 'lat',
'fips_cum_cases',
'white_pct', 'black_pct',
'hispanic_pct',
'age65andolder_pct',
'lesshs_pct', 'rural_pct',
'ruralurban_cc',
'state_popn',
'POPESTIMATE2019',
'lo_trump_vote_16',
'fips_n_cases_per1k',
'fips_n_cases_per1k_log',
'fips_n_cases_lo',...
'fips_lag1_n_cases_per1k_log',
'fips_lag1_n_cases_lo',
'fips_n_deaths_per1k',
'fips_n_deaths_per1k_log',
'fips_n_deaths_lo',
'fips_lag1_n_deaths_per1k',
'fips_lag1_n_deaths_per1k_log',
'fips_lag1_n_deaths_lo',
'state_n_cases_per1k',
'state_n_cases_per1k_log',
'state_n_cases_lo',
'state_lag1_n_cases_per1k', ...])])),
('lasso', Lasso(alpha=0.0071968567300115215, random_state=0))])ColumnTransformer(transformers=[('scale', StandardScaler(),
['week_num', 'lon', 'lat', 'fips_cum_cases',
'white_pct', 'black_pct', 'hispanic_pct',
'age65andolder_pct', 'lesshs_pct',
'rural_pct', 'ruralurban_cc', 'state_popn',
'POPESTIMATE2019', 'lo_trump_vote_16',
'fips_n_cases_per1k',
'fips_n_cases_per1k_log', 'fips_n_cases_lo',
'fips_lag1_n_cases_per1k',
'fips_lag1_n_cases_per1k_log',
'fips_lag1_n_cases_lo', 'fips_n_deaths_per1k',
'fips_n_deaths_per1k_log', 'fips_n_deaths_lo',
'fips_lag1_n_deaths_per1k',
'fips_lag1_n_deaths_per1k_log',
'fips_lag1_n_deaths_lo',
'state_n_cases_per1k',
'state_n_cases_per1k_log', 'state_n_cases_lo',
'state_lag1_n_cases_per1k', ...])])['week_num', 'lon', 'lat', 'fips_cum_cases', 'white_pct', 'black_pct', 'hispanic_pct', 'age65andolder_pct', 'lesshs_pct', 'rural_pct', 'ruralurban_cc', 'state_popn', 'POPESTIMATE2019', 'lo_trump_vote_16', 'fips_n_cases_per1k', 'fips_n_cases_per1k_log', 'fips_n_cases_lo', 'fips_lag1_n_cases_per1k', 'fips_lag1_n_cases_per1k_log', 'fips_lag1_n_cases_lo', 'fips_n_deaths_per1k', 'fips_n_deaths_per1k_log', 'fips_n_deaths_lo', 'fips_lag1_n_deaths_per1k', 'fips_lag1_n_deaths_per1k_log', 'fips_lag1_n_deaths_lo', 'state_n_cases_per1k', 'state_n_cases_per1k_log', 'state_n_cases_lo', 'state_lag1_n_cases_per1k', 'state_lag1_n_cases_per1k_log', 'state_lag1_n_cases_lo', 'state_n_deaths_per1k', 'state_n_deaths_per1k_log', 'state_n_deaths_lo', 'state_lag1_n_deaths_per1k', 'state_lag1_n_deaths_per1k_log', 'state_lag1_n_deaths_lo', 'country_n_cases_per1k', 'country_n_cases_per1k_log', 'country_n_cases_lo', 'country_lag1_n_cases_per1k', 'country_lag1_n_cases_per1k_log', 'country_lag1_n_cases_lo', 'country_n_deaths_per1k', 'country_n_deaths_per1k_log', 'country_n_deaths_lo', 'country_lag1_n_deaths_per1k', 'country_lag1_n_deaths_per1k_log', 'country_lag1_n_deaths_lo', 'predrt_0', 'predrt_3', 'predrt_12', 'log_pop', 'log_rank', 'lo_trump_vote']
StandardScaler()
Lasso(alpha=0.0071968567300115215, random_state=0)
lasso_results = pd.DataFrame({"feature_names" : mod[-2].get_feature_names_out(),
"coefs" : mod[-1].coef_
})
lasso_results.sort_values("coefs", ascending=False)
| feature_names | coefs | |
|---|---|---|
| 46 | scale__country_n_deaths_lo | 0.184288 |
| 54 | scale__log_rank | 0.127895 |
| 28 | scale__state_n_cases_lo | 0.054070 |
| 26 | scale__state_n_cases_per1k | 0.011341 |
| 34 | scale__state_n_deaths_lo | 0.010528 |
| 8 | scale__lesshs_pct | 0.006633 |
| 40 | scale__country_n_cases_lo | 0.000000 |
| 31 | scale__state_lag1_n_cases_lo | 0.000000 |
| 32 | scale__state_n_deaths_per1k | 0.000000 |
| 33 | scale__state_n_deaths_per1k_log | 0.000000 |
| 35 | scale__state_lag1_n_deaths_per1k | 0.000000 |
| 36 | scale__state_lag1_n_deaths_per1k_log | -0.000000 |
| 37 | scale__state_lag1_n_deaths_lo | 0.000000 |
| 38 | scale__country_n_cases_per1k | 0.000000 |
| 39 | scale__country_n_cases_per1k_log | 0.000000 |
| 42 | scale__country_lag1_n_cases_per1k_log | 0.000000 |
| 41 | scale__country_lag1_n_cases_per1k | -0.000000 |
| 29 | scale__state_lag1_n_cases_per1k | 0.000000 |
| 43 | scale__country_lag1_n_cases_lo | 0.000000 |
| 44 | scale__country_n_deaths_per1k | 0.000000 |
| 45 | scale__country_n_deaths_per1k_log | 0.000000 |
| 47 | scale__country_lag1_n_deaths_per1k | -0.000000 |
| 48 | scale__country_lag1_n_deaths_per1k_log | -0.000000 |
| 49 | scale__country_lag1_n_deaths_lo | -0.000000 |
| 50 | scale__predrt_0 | 0.000000 |
| 52 | scale__predrt_12 | -0.000000 |
| 53 | scale__log_pop | 0.000000 |
| 30 | scale__state_lag1_n_cases_per1k_log | 0.000000 |
| 55 | scale__lo_trump_vote | -0.000000 |
| 14 | scale__fips_n_cases_per1k | 0.000000 |
| 18 | scale__fips_lag1_n_cases_per1k_log | -0.000000 |
| 10 | scale__ruralurban_cc | 0.000000 |
| 12 | scale__POPESTIMATE2019 | 0.000000 |
| 2 | scale__lat | 0.000000 |
| 27 | scale__state_n_cases_per1k_log | 0.000000 |
| 15 | scale__fips_n_cases_per1k_log | 0.000000 |
| 16 | scale__fips_n_cases_lo | -0.000000 |
| 6 | scale__hispanic_pct | -0.000000 |
| 17 | scale__fips_lag1_n_cases_per1k | -0.000000 |
| 19 | scale__fips_lag1_n_cases_lo | -0.000000 |
| 21 | scale__fips_n_deaths_per1k_log | 0.000000 |
| 22 | scale__fips_n_deaths_lo | -0.000000 |
| 24 | scale__fips_lag1_n_deaths_per1k_log | -0.000000 |
| 25 | scale__fips_lag1_n_deaths_lo | -0.000000 |
| 7 | scale__age65andolder_pct | -0.000000 |
| 20 | scale__fips_n_deaths_per1k | -0.003268 |
| 23 | scale__fips_lag1_n_deaths_per1k | -0.012095 |
| 3 | scale__fips_cum_cases | -0.020631 |
| 51 | scale__predrt_3 | -0.031312 |
| 1 | scale__lon | -0.032317 |
| 4 | scale__white_pct | -0.038656 |
| 9 | scale__rural_pct | -0.048886 |
| 11 | scale__state_popn | -0.051594 |
| 5 | scale__black_pct | -0.110805 |
| 13 | scale__lo_trump_vote_16 | -0.148217 |
| 0 | scale__week_num | -0.456441 |
from pygam import LinearGAM, s, f
gam = LinearGAM(s(0) + s(1)).fit(covid_df[['week_num', 'country_n_deaths_per1k']], covid_df.log_odds_of_covid_article)
gam.summary()
LinearGAM
=============================================== ==========================================================
Distribution: NormalDist Effective DoF: 30.9397
Link Function: IdentityLink Log Likelihood: -16531.9023
Number of Samples: 13458 AIC: 33127.6838
AICc: 33127.8406
GCV: 0.5541
Scale: 0.5518
Pseudo R-Squared: 0.2389
==========================================================================================================
Feature Function Lambda Rank EDoF P > x Sig. Code
================================= ==================== ============ ============ ============ ============
s(0) [0.6] 20 19.0 1.11e-16 ***
s(1) [0.6] 20 11.9 5.18e-01
intercept 1 0.0 9.23e-02 .
==========================================================================================================
Significance codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
WARNING: Fitting splines and a linear function to a feature introduces a model identifiability problem
which can cause p-values to appear significant when they are not.
WARNING: p-values calculated in this manner behave correctly for un-penalized models or models with
known smoothing parameters, but when smoothing parameters have been estimated, the p-values
are typically lower than they should be, meaning that the tests reject the null too readily.
/var/folders/rf/4jz25sh1733_8b28phhhrlfh0000gn/T/ipykernel_12153/3358381670.py:1: UserWarning: KNOWN BUG: p-values computed in this summary are likely much smaller than they should be. Please do not make inferences based on these values! Collaborate on a solution, and stay up to date at: github.com/dswah/pyGAM/issues/163
for i, term in enumerate(gam.terms):
if term.isintercept:
continue
XX = gam.generate_X_grid(term=i)
pdep, confi = gam.partial_dependence(term=i, X=XX, width=0.95)
plt.figure()
plt.plot(XX[:, term.feature], pdep)
plt.plot(XX[:, term.feature], confi, c='r', ls='--')
plt.title(repr(term))
plt.show()
averaged_data = covid_df.groupby("date").agg(
cases = pd.NamedAgg("country_n_cases_per1k","first"),
deaths = pd.NamedAgg("country_n_deaths_per1k","first"),
article_odds = pd.NamedAgg("log_odds_of_covid_article","mean")
).reset_index()
fig = px.line(pd.melt(averaged_data,id_vars="date"),x="date",y="value",facet_col="variable")
fig.update_yaxes(matches=None)
fig.show()